Also known as a Cophylo Plot or Co-phylogeny Plot.
A Tanglegram is a visualisation that consists of two Dendrogram trees displayed side-by-side that share the same set of leaves. Connection lines are drawn between these leaves to show the matches between the two trees. As a visualisation method, Tanglegrams are often implemented to compare and display the concordance (similarity of traits) between two datasets of hierarchical clustering.
Tanglegrams are particularly useful in the field of biology, for example, for comparing evolutionary relationships, especially in detecting the coevolutionary processes between two (or more) taxa (co-phylogeny).
Links to tools or code that can draw a Tanglegram:
- Dendroscope
- Jane 4
- Mesquite
- The Python Package Index (Python)
- Rdocumentation: cophyloplot (R)
- R Graph Gallery (R)
Examples of a Tanglegram
Cophylogeny (tanglegram) showing maximum-likelihood trees estimated with IQTree from 189 low-copy nuclear loci generated in this study (A) and from traditional markers (B).
A customised target capture sequencing tool for molecular identification of Aloe vera and relatives — ResearchGate
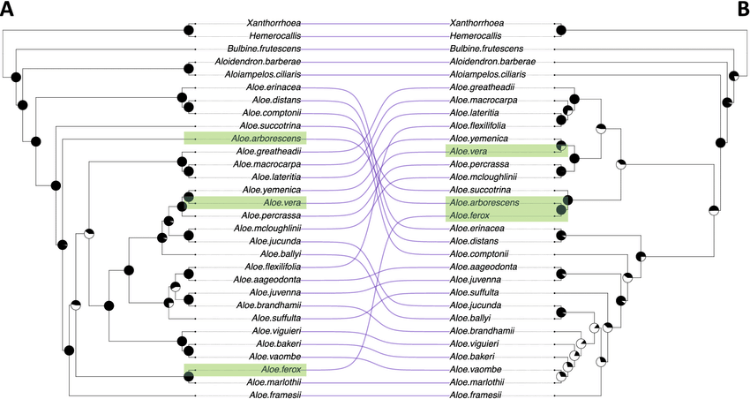
A tanglegram between a phylogeny of sections of Ficus (a) and that of their associated genera of pollinating wasps (b). Adapted from (Machado et al., 2005).
Tanglegrams for rooted phylogenetic trees and networks — ResearchGate
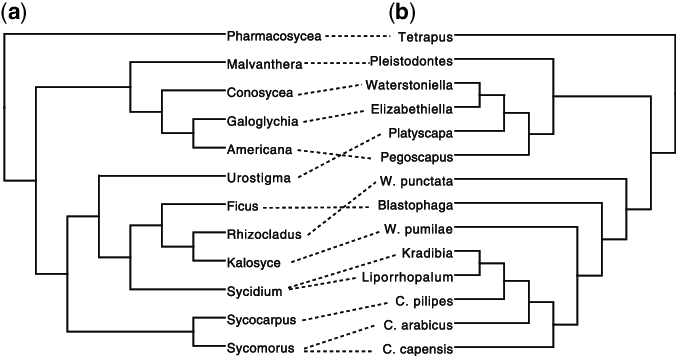
Tanglegram of cophylogenetic relationships between hosts (left) and Wolbachia strains (right).
Supergroup F Wolbachia in terrestrial isopods: Horizontal transmission from termites? — ResearchGate
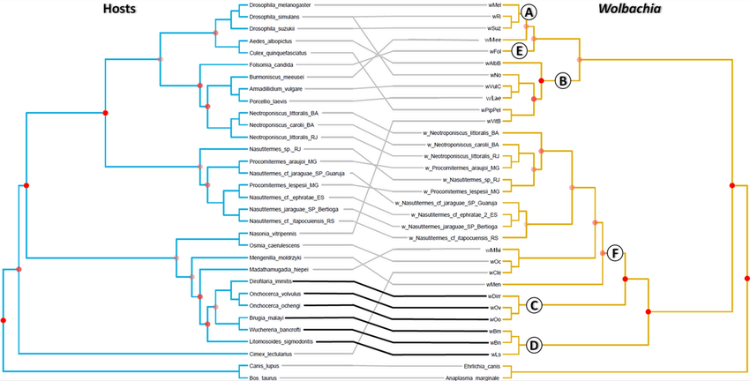
Fig. 6: The tanglegrams of gyrodactylids and host fishes inferred using the P18SHMITO-ML dataset. The figure contains a parasite tree (on the left), a host tree (on the right) with the order-level annotation, and a set of host-parasite associations (the host range of each parasite).
Geography, phylogeny and host switch drive the coevolution of parasitic Gyrodactylus flatworms and their hosts — Parasites & Vectors

A tanglegram of waterbirds and louse phylogenies based on Van Tuinen et al. (2001) and Smith et al. (2004), respectively.
Reinterpreting the origins of flamingo lice: Cospeciation or host-switching?
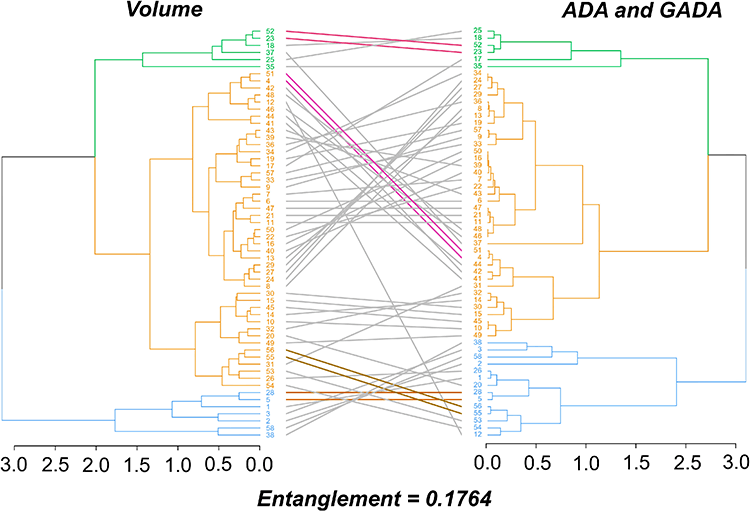
Figure 6. Tanglegram comparing dendrograms between volume and site index by the ADA and GADA approach, based on hierarchical clustering, in clonal teak (Tectona grandis Linn F.) plantations in the Eastern Brazilian Amazon. Colored lines connect common branches between the two dendrograms.
Approaches to Forest Site Classification as an Indicator of Teak Volume Production — MDPI
Related posts:
Chart Snapshot: Dendrograms
Chart Snapshot: Circular Dendrograms
